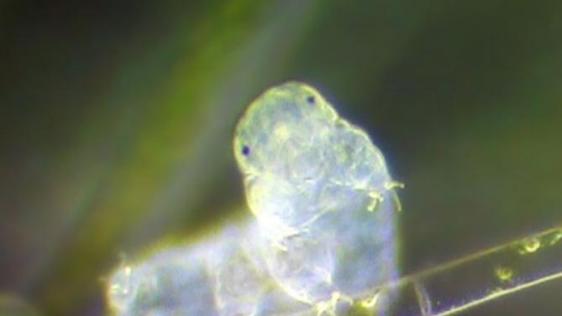
Washington (AFP) – The eight-legged water bear — a hardy, nearly microscopic animal resembling its mammal namesake — gets a huge chunk of its DNA from foreign organisms such as bacteria and plants, scientists have revealed.
These genes, the researchers suggest, help the tiny animals, also known as moss piglets or tardigrades, survive in the harshest of environments.
Water bears, which live all over the world, are usually 0.020 inches (0.5 millimetres) long and move very slowly and clumsily on their multitude of legs.
These highly adaptable creatures can survive extreme temperatures.
Even after being stuck in a freezer at -112 degrees Fahrenheit (-80 Celsius) for 10 years, they can start moving around again about 20 minutes after thawing.
By sequencing these creatures’ genome, researchers from the University of North Carolina (UNC) at Chapel Hill were surprised to find that 17.5 percent — nearly a sixth — of the genome came from foreign organisms.
For most animals, less than one percent of their genome comes from foreign DNA.
The microscopic rotifer previously held the record, with eight percent of its genome coming from foreign DNA.
“We had no idea that an animal genome could be composed of so much foreign DNA,” said co-author Bob Goldstein of UNC’s College of Arts and Sciences.
“We knew many animals acquire foreign genes, but we had no idea that it happens to this degree.”
– New insight on evolution –
The study, published in Monday’s edition of the Proceedings of the National Academy of Sciences, also made unusual findings about how DNA is inherited.
Goldstein, first author Thomas Boothby and colleagues found that water bears obtain about 6,000 foreign genes mostly from bacteria, as well as plants, fungi and Archaea single-cell organisms.
“Animals that can survive extreme stresses may be particularly prone to acquiring foreign genes — and bacterial genes might be better able to withstand stresses than animal ones,” said Boothby, a postdoctoral fellow in Goldstein’s lab.
Indeed, bacteria have survived the most extreme environments on Earth for billions of years.
Water bears acquire foreign genes through horizontal gene transfer, a process by which species swap genetic material instead of inheriting DNA from parents.
“With horizontal gene transfer becoming more widely accepted and more well-known, at least in certain organisms, it is beginning to change the way we think about evolution and inheritance of genetic material and the stability of genomes,” said Boothby.
Researchers said the DNA likely gets inside the genome randomly but what remains allows water bears to survive in the most hostile environments.
Under intense stress, such as extreme dryness, the water bear’s DNA breaks up into small pieces, according to the research team.
Once the cell rehydrates, its membrane and nucleus housing the DNA temporarily becomes leaky and allows other large molecules to pass through easily.
They thus repair their own damaged DNA while also absorbing foreign DNA as the cell rehydrates, forming a patchworks of genes from different species.
“So instead of thinking of the tree of life, we can think about the web of life and genetic material crossing from branch to branch,” Boothby explained.
“So it’s exciting. We are beginning to adjust our understanding of how evolution works.”
Source: Tiny water bears are huge DNA thieves: study – Yahoo News